In 2021, Annogen, the Amsterdam-based biotech company behind the SuRE technology for the functional annotation of the non-coding part of the genome, and KeyGene started a collaboration to systematically screen for variants that result in important crop traits.
This project has now led to the identification of sequence variants that upregulate important traits in plants. More generally the results demonstrate the efficacy of SuRE-based sequence variant screening in plants for the improvement of crop traits such as higher yields and resistance to drought and pathogens.
In the project, KeyGene provided Annogen with several hundred candidate sequence variants occurring in promoter or enhancer regions of trait-relevant genes. These natural variants came from diverse tomato types. SuRE libraries for these variants were generated by Annogen and analyzed in tomato protoplasts. This allowed for the identification of several candidate variants which positively change the expression of trait-relevant genes. Two of these candidates were subsequently tested in planta in a homozygous setting.
Prediction and results
The first variant was predicted to increase the expression of a gene that plays a critical role in biotic resistance, and indeed, a ~5-fold increase could be confirmed by qPCR after infection.
The second variant was expected to increase the expression of a gene involved in the response to wounding. Indeed a ~2-fold upregulation after leaf damage of this gene was observed.
"The experiments demonstrate the value of SuRE in identifying promising gain-of-function sequence variants in a massively parallel setting, without the need to grow individual plants," said Paul Bundock, senior scientist at KeyGene.
In crops
SuRE can be applied to any crop and trait and is being further optimized at Annogen for in vivo applications using Agrobacterium. "We are very excited that our SuRE approach identified sequence variants that in plants led to increased expression of trait-relevant genes. These results even exceed what we hoped for when we started this project 3 years ago" said Joris van Arensbergen, Annogen's founder and CEO.
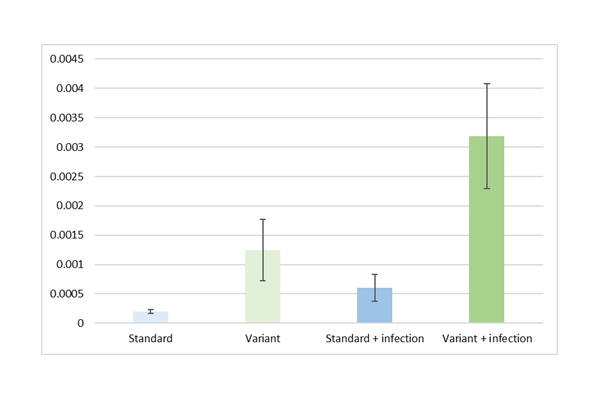
Graph: Thanks to the use of Annogen's SuRE platform and KeyGene's insights into crop genetics, KeyGene, and Annogen were able to identify plants with an increased expression of genes involved in some important crop traits.
Source: KeyGene
